Specificity¶
Module Interface¶
- class torchmetrics.Specificity(task: Literal['binary', 'multiclass', 'multilabel'], threshold: float = 0.5, num_classes: Optional[int] = None, num_labels: Optional[int] = None, average: Optional[Literal['micro', 'macro', 'weighted', 'none']] = 'micro', multidim_average: Optional[Literal['global', 'samplewise']] = 'global', top_k: Optional[int] = 1, ignore_index: Optional[int] = None, validate_args: bool = True, **kwargs: Any)[source]
Computes Specificity.
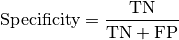
Where
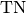 and
and 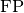 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.This function is a simple wrapper to get the task specific versions of this metric, which is done by setting the
taskargument to either'binary','multiclass'ormultilabel. See the documentation ofBinarySpecificity,MulticlassSpecificityandMultilabelSpecificityfor the specific details of each argument influence and examples.- Legacy Example:
>>> preds = torch.tensor([2, 0, 2, 1]) >>> target = torch.tensor([1, 1, 2, 0]) >>> specificity = Specificity(task="multiclass", average='macro', num_classes=3) >>> specificity(preds, target) tensor(0.6111) >>> specificity = Specificity(task="multiclass", average='micro', num_classes=3) >>> specificity(preds, target) tensor(0.6250)
BinarySpecificity¶
- class torchmetrics.classification.BinarySpecificity(threshold=0.5, multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes Specificity for binary tasks:
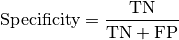
Where
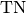 and
and 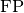 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds(int or float tensor):(N, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary {0,1} predictionsmultidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal, the metric returns a scalar value. Ifmultidim_averageis set tosamplewise, the metric returns(N,)vector consisting of a scalar value per sample.
- Example (preds is int tensor):
>>> from torchmetrics.classification import BinarySpecificity >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0, 0, 1, 1, 0, 1]) >>> metric = BinarySpecificity() >>> metric(preds, target) tensor(0.6667)
- Example (preds is float tensor):
>>> from torchmetrics.classification import BinarySpecificity >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0.11, 0.22, 0.84, 0.73, 0.33, 0.92]) >>> metric = BinarySpecificity() >>> metric(preds, target) tensor(0.6667)
- Example (multidim tensors):
>>> from torchmetrics.classification import BinarySpecificity >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> metric = BinarySpecificity(multidim_average='samplewise') >>> metric(preds, target) tensor([0.0000, 0.3333])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
- compute()[source]
Computes the final statistics.
- Return type
- Returns
The metric returns a tensor of shape
(..., 5), where the last dimension corresponds to[tp, fp, tn, fn, sup](supstands for support and equalstp + fn). The shape depends on themultidim_averageparameter:If
multidim_averageis set toglobal, the shape will be(5,)If
multidim_averageis set tosamplewise, the shape will be(N, 5)
MulticlassSpecificity¶
- class torchmetrics.classification.MulticlassSpecificity(num_classes, top_k=1, average='macro', multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes Specificity for multiclass tasks:
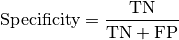
Where
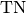 and
and 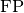 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds:(N, ...)(int tensor) or(N, C, ..)(float tensor). If preds is a floating point we applytorch.argmaxalong theCdimension to automatically convert probabilities/logits into an int tensor.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
num_classes¶ (
int) – Integer specifing the number of classesaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
top_k¶ (
int) – Number of highest probability or logit score predictions considered to find the correct label. Only works whenpredscontain probabilities/logits.multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.classification import MulticlassSpecificity >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([2, 1, 0, 1]) >>> metric = MulticlassSpecificity(num_classes=3) >>> metric(preds, target) tensor(0.8889) >>> metric = MulticlassSpecificity(num_classes=3, average=None) >>> metric(preds, target) tensor([1.0000, 0.6667, 1.0000])
- Example (preds is float tensor):
>>> from torchmetrics.classification import MulticlassSpecificity >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([ ... [0.16, 0.26, 0.58], ... [0.22, 0.61, 0.17], ... [0.71, 0.09, 0.20], ... [0.05, 0.82, 0.13], ... ]) >>> metric = MulticlassSpecificity(num_classes=3) >>> metric(preds, target) tensor(0.8889) >>> metric = MulticlassSpecificity(num_classes=3, average=None) >>> metric(preds, target) tensor([1.0000, 0.6667, 1.0000])
- Example (multidim tensors):
>>> from torchmetrics.classification import MulticlassSpecificity >>> target = torch.tensor([[[0, 1], [2, 1], [0, 2]], [[1, 1], [2, 0], [1, 2]]]) >>> preds = torch.tensor([[[0, 2], [2, 0], [0, 1]], [[2, 2], [2, 1], [1, 0]]]) >>> metric = MulticlassSpecificity(num_classes=3, multidim_average='samplewise') >>> metric(preds, target) tensor([0.7500, 0.6556]) >>> metric = MulticlassSpecificity(num_classes=3, multidim_average='samplewise', average=None) >>> metric(preds, target) tensor([[0.7500, 0.7500, 0.7500], [0.8000, 0.6667, 0.5000]])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
- compute()[source]
Computes the final statistics.
- Return type
- Returns
The metric returns a tensor of shape
(..., 5), where the last dimension corresponds to[tp, fp, tn, fn, sup](supstands for support and equalstp + fn). The shape depends onaverageandmultidim_averageparameters:If
multidim_averageis set toglobalIf
average='micro'/'macro'/'weighted', the shape will be(5,)If
average=None/'none', the shape will be(C, 5)If
multidim_averageis set tosamplewiseIf
average='micro'/'macro'/'weighted', the shape will be(N, 5)If
average=None/'none', the shape will be(N, C, 5)
MultilabelSpecificity¶
- class torchmetrics.classification.MultilabelSpecificity(num_labels, threshold=0.5, average='macro', multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes Specificity for multilabel tasks.
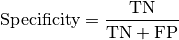
Where
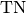 and
and 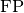 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds(int or float tensor):(N, C, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, C, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary (0,1) predictionsaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal- Ifaverage='micro'/'macro'/'weighted', the output will be a scalar tensor - Ifaverage=None/'none', the shape will be(C,)If
multidim_averageis set tosamplewise- Ifaverage='micro'/'macro'/'weighted', the shape will be(N,)- Ifaverage=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.classification import MultilabelSpecificity >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0, 0, 1], [1, 0, 1]]) >>> metric = MultilabelSpecificity(num_labels=3) >>> metric(preds, target) tensor(0.6667) >>> metric = MultilabelSpecificity(num_labels=3, average=None) >>> metric(preds, target) tensor([1., 1., 0.])
- Example (preds is float tensor):
>>> from torchmetrics.classification import MultilabelSpecificity >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0.11, 0.22, 0.84], [0.73, 0.33, 0.92]]) >>> metric = MultilabelSpecificity(num_labels=3) >>> metric(preds, target) tensor(0.6667) >>> metric = MultilabelSpecificity(num_labels=3, average=None) >>> metric(preds, target) tensor([1., 1., 0.])
- Example (multidim tensors):
>>> from torchmetrics.classification import MultilabelSpecificity >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> metric = MultilabelSpecificity(num_labels=3, multidim_average='samplewise') >>> metric(preds, target) tensor([0.0000, 0.3333]) >>> metric = MultilabelSpecificity(num_labels=3, multidim_average='samplewise', average=None) >>> metric(preds, target) tensor([[0., 0., 0.], [0., 0., 1.]])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
- compute()[source]
Computes the final statistics.
- Return type
- Returns
The metric returns a tensor of shape
(..., 5), where the last dimension corresponds to[tp, fp, tn, fn, sup](supstands for support and equalstp + fn). The shape depends onaverageandmultidim_averageparameters:If
multidim_averageis set toglobalIf
average='micro'/'macro'/'weighted', the shape will be(5,)If
average=None/'none', the shape will be(C, 5)If
multidim_averageis set tosamplewiseIf
average='micro'/'macro'/'weighted', the shape will be(N, 5)If
average=None/'none', the shape will be(N, C, 5)
Functional Interface¶
- torchmetrics.functional.specificity(preds, target, task, threshold=0.5, num_classes=None, num_labels=None, average='micro', multidim_average='global', top_k=1, ignore_index=None, validate_args=True)[source]
Computes Specificity.
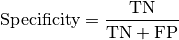
Where
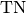 and
and 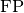 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.This function is a simple wrapper to get the task specific versions of this metric, which is done by setting the
taskargument to either'binary','multiclass'ormultilabel. See the documentation ofbinary_specificity(),multiclass_specificity()andmultilabel_specificity()for the specific details of each argument influence and examples.- LegacyExample:
>>> preds = torch.tensor([2, 0, 2, 1]) >>> target = torch.tensor([1, 1, 2, 0]) >>> specificity(preds, target, task="multiclass", average='macro', num_classes=3) tensor(0.6111) >>> specificity(preds, target, task="multiclass", average='micro', num_classes=3) tensor(0.6250)
- Return type
binary_specificity¶
- torchmetrics.functional.classification.binary_specificity(preds, target, threshold=0.5, multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes Specificity for binary tasks:
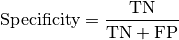
Where
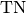 and
and 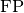 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds(int or float tensor):(N, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary {0,1} predictionsmultidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Return type
- Returns
If
multidim_averageis set toglobal, the metric returns a scalar value. Ifmultidim_averageis set tosamplewise, the metric returns(N,)vector consisting of a scalar value per sample.
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import binary_specificity >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0, 0, 1, 1, 0, 1]) >>> binary_specificity(preds, target) tensor(0.6667)
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import binary_specificity >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0.11, 0.22, 0.84, 0.73, 0.33, 0.92]) >>> binary_specificity(preds, target) tensor(0.6667)
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import binary_specificity >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> binary_specificity(preds, target, multidim_average='samplewise') tensor([0.0000, 0.3333])
multiclass_specificity¶
- torchmetrics.functional.classification.multiclass_specificity(preds, target, num_classes, average='macro', top_k=1, multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes Specificity for multiclass tasks:
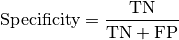
Where
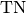 and
and 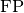 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds:(N, ...)(int tensor) or(N, C, ..)(float tensor). If preds is a floating point we applytorch.argmaxalong theCdimension to automatically convert probabilities/logits into an int tensor.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
num_classes¶ (
int) – Integer specifing the number of classesaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
top_k¶ (
int) – Number of highest probability or logit score predictions considered to find the correct label. Only works whenpredscontain probabilities/logits.multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import multiclass_specificity >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([2, 1, 0, 1]) >>> multiclass_specificity(preds, target, num_classes=3) tensor(0.8889) >>> multiclass_specificity(preds, target, num_classes=3, average=None) tensor([1.0000, 0.6667, 1.0000])
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import multiclass_specificity >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([ ... [0.16, 0.26, 0.58], ... [0.22, 0.61, 0.17], ... [0.71, 0.09, 0.20], ... [0.05, 0.82, 0.13], ... ]) >>> multiclass_specificity(preds, target, num_classes=3) tensor(0.8889) >>> multiclass_specificity(preds, target, num_classes=3, average=None) tensor([1.0000, 0.6667, 1.0000])
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import multiclass_specificity >>> target = torch.tensor([[[0, 1], [2, 1], [0, 2]], [[1, 1], [2, 0], [1, 2]]]) >>> preds = torch.tensor([[[0, 2], [2, 0], [0, 1]], [[2, 2], [2, 1], [1, 0]]]) >>> multiclass_specificity(preds, target, num_classes=3, multidim_average='samplewise') tensor([0.7500, 0.6556]) >>> multiclass_specificity(preds, target, num_classes=3, multidim_average='samplewise', average=None) tensor([[0.7500, 0.7500, 0.7500], [0.8000, 0.6667, 0.5000]])
multilabel_specificity¶
- torchmetrics.functional.classification.multilabel_specificity(preds, target, num_labels, threshold=0.5, average='macro', multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes Specificity for multilabel tasks.
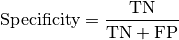
Where
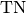 and
and 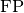 represent the number of true negatives and
false positives respecitively.
represent the number of true negatives and
false positives respecitively.Accepts the following input tensors:
preds(int or float tensor):(N, C, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, C, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary (0,1) predictionsaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import multilabel_specificity >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0, 0, 1], [1, 0, 1]]) >>> multilabel_specificity(preds, target, num_labels=3) tensor(0.6667) >>> multilabel_specificity(preds, target, num_labels=3, average=None) tensor([1., 1., 0.])
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import multilabel_specificity >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0.11, 0.22, 0.84], [0.73, 0.33, 0.92]]) >>> multilabel_specificity(preds, target, num_labels=3) tensor(0.6667) >>> multilabel_specificity(preds, target, num_labels=3, average=None) tensor([1., 1., 0.])
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import multilabel_specificity >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> multilabel_specificity(preds, target, num_labels=3, multidim_average='samplewise') tensor([0.0000, 0.3333]) >>> multilabel_specificity(preds, target, num_labels=3, multidim_average='samplewise', average=None) tensor([[0., 0., 0.], [0., 0., 1.]])
