F-1 Score¶
Module Interface¶
F1Score¶
- class torchmetrics.F1Score(num_classes=None, threshold=0.5, average='micro', mdmc_average=None, ignore_index=None, top_k=None, multiclass=None, **kwargs)[source]
F1 Score.
Note
From v0.10 an
'binary_*','multiclass_*','multilabel_*'version now exist of each classification metric. Moving forward we recommend using these versions. This base metric will still work as it did prior to v0.10 until v0.11. From v0.11 the task argument introduced in this metric will be required and the general order of arguments may change, such that this metric will just function as an single entrypoint to calling the three specialized versions.Computes F1 metric.
F1 metrics correspond to a harmonic mean of the precision and recall scores. Works with binary, multiclass, and multilabel data. Accepts logits or probabilities from a model output or integer class values in prediction. Works with multi-dimensional preds and target.
Forward accepts
preds(float or long tensor):(N, ...)or(N, C, ...)where C is the number of classestarget(long tensor):(N, ...)
If preds and target are the same shape and preds is a float tensor, we use the
self.thresholdargument. This is the case for binary and multi-label logits.If preds has an extra dimension as in the case of multi-class scores we perform an argmax on
dim=1.- Parameters
num_classes¶ (
Optional[int]) – Number of classes. Necessary for'macro','weighted'andNoneaverage methods.threshold¶ (
float) – Threshold for transforming probability or logit predictions to binary(0,1)predictions, in the case of binary or multi-label inputs. Default value of0.5corresponds to input being probabilities.average¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied. Should be one of the following:
'micro'[default]: Calculate the metric globally, across all samples and classes.'macro': Calculate the metric for each class separately, and average the metrics across classes (with equal weights for each class).'weighted': Calculate the metric for each class separately, and average the metrics across classes, weighting each class by its support (tp + fn).'none'orNone: Calculate the metric for each class separately, and return the metric for every class.'samples': Calculate the metric for each sample, and average the metrics across samples (with equal weights for each sample).
Note
What is considered a sample in the multi-dimensional multi-class case depends on the value of
mdmc_average.mdmc_average¶ (
Optional[str]) –Defines how averaging is done for multi-dimensional multi-class inputs (on top of the
averageparameter). Should be one of the following:None[default]: Should be left unchanged if your data is not multi-dimensional multi-class.'samplewise': In this case, the statistics are computed separately for each sample on theNaxis, and then averaged over samples. The computation for each sample is done by treating the flattened extra axes...(see Input types) as theNdimension within the sample, and computing the metric for the sample based on that.'global': In this case theNand...dimensions of the inputs (see Input types) are flattened into a newN_Xsample axis, i.e. the inputs are treated as if they were(N_X, C). From here on theaverageparameter applies as usual.
ignore_index¶ (
Optional[int]) – Integer specifying a target class to ignore. If given, this class index does not contribute to the returned score, regardless of reduction method. If an index is ignored, andaverage=Noneor'none', the score for the ignored class will be returned asnan.top_k¶ (
Optional[int]) – Number of the highest probability or logit score predictions considered finding the correct label, relevant only for (multi-dimensional) multi-class inputs. The default value (None) will be interpreted as 1 for these inputs. Should be left at default (None) for all other types of inputs.multiclass¶ (
Optional[bool]) – Used only in certain special cases, where you want to treat inputs as a different type than what they appear to be. See the parameter’s documentation section for a more detailed explanation and examples.kwargs¶ (
Any) – Additional keyword arguments, see Advanced metric settings for more info.
Example
>>> import torch >>> from torchmetrics import F1Score >>> target = torch.tensor([0, 1, 2, 0, 1, 2]) >>> preds = torch.tensor([0, 2, 1, 0, 0, 1]) >>> f1 = F1Score(num_classes=3) >>> f1(preds, target) tensor(0.3333)
Initializes internal Module state, shared by both nn.Module and ScriptModule.
BinaryF1Score¶
- class torchmetrics.classification.BinaryF1Score(threshold=0.5, multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes F-1 score for binary tasks:
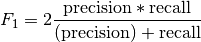
Accepts the following input tensors:
preds(int or float tensor):(N, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary {0,1} predictionsmultidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal, the metric returns a scalar value. Ifmultidim_averageis set tosamplewise, the metric returns(N,)vector consisting of a scalar value per sample.
- Example (preds is int tensor):
>>> from torchmetrics.classification import BinaryF1Score >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0, 0, 1, 1, 0, 1]) >>> metric = BinaryF1Score() >>> metric(preds, target) tensor(0.6667)
- Example (preds is float tensor):
>>> from torchmetrics.classification import BinaryF1Score >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0.11, 0.22, 0.84, 0.73, 0.33, 0.92]) >>> metric = BinaryF1Score() >>> metric(preds, target) tensor(0.6667)
- Example (multidim tensors):
>>> from torchmetrics.classification import BinaryF1Score >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> metric = BinaryF1Score(multidim_average='samplewise') >>> metric(preds, target) tensor([0.5000, 0.0000])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
MulticlassF1Score¶
- class torchmetrics.classification.MulticlassF1Score(num_classes, top_k=1, average='macro', multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes F-1 score for multiclass tasks:
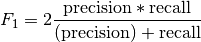
Accepts the following input tensors:
preds:(N, ...)(int tensor) or(N, C, ..)(float tensor). If preds is a floating point we applytorch.argmaxalong theCdimension to automatically convert probabilities/logits into an int tensor.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
preds¶ – Tensor with predictions
target¶ – Tensor with true labels
num_classes¶ (
int) – Integer specifing the number of classesaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
top_k¶ (
int) – Number of highest probability or logit score predictions considered to find the correct label. Only works whenpredscontain probabilities/logits.multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.classification import MulticlassF1Score >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([2, 1, 0, 1]) >>> metric = MulticlassF1Score(num_classes=3) >>> metric(preds, target) tensor(0.7778) >>> metric = MulticlassF1Score(num_classes=3, average=None) >>> metric(preds, target) tensor([0.6667, 0.6667, 1.0000])
- Example (preds is float tensor):
>>> from torchmetrics.classification import MulticlassF1Score >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([ ... [0.16, 0.26, 0.58], ... [0.22, 0.61, 0.17], ... [0.71, 0.09, 0.20], ... [0.05, 0.82, 0.13], ... ]) >>> metric = MulticlassF1Score(num_classes=3) >>> metric(preds, target) tensor(0.7778) >>> metric = MulticlassF1Score(num_classes=3, average=None) >>> metric(preds, target) tensor([0.6667, 0.6667, 1.0000])
- Example (multidim tensors):
>>> from torchmetrics.classification import MulticlassF1Score >>> target = torch.tensor([[[0, 1], [2, 1], [0, 2]], [[1, 1], [2, 0], [1, 2]]]) >>> preds = torch.tensor([[[0, 2], [2, 0], [0, 1]], [[2, 2], [2, 1], [1, 0]]]) >>> metric = MulticlassF1Score(num_classes=3, multidim_average='samplewise') >>> metric(preds, target) tensor([0.4333, 0.2667]) >>> metric = MulticlassF1Score(num_classes=3, multidim_average='samplewise', average=None) >>> metric(preds, target) tensor([[0.8000, 0.0000, 0.5000], [0.0000, 0.4000, 0.4000]])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
MultilabelF1Score¶
- class torchmetrics.classification.MultilabelF1Score(num_labels, threshold=0.5, average='macro', multidim_average='global', ignore_index=None, validate_args=True, **kwargs)[source]
Computes F-1 score for multilabel tasks:
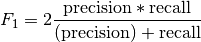
Accepts the following input tensors:
preds(int or float tensor):(N, C, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, C, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary (0,1) predictionsaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)`
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.classification import MultilabelF1Score >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0, 0, 1], [1, 0, 1]]) >>> metric = MultilabelF1Score(num_labels=3) >>> metric(preds, target) tensor(0.5556) >>> metric = MultilabelF1Score(num_labels=3, average=None) >>> metric(preds, target) tensor([1.0000, 0.0000, 0.6667])
- Example (preds is float tensor):
>>> from torchmetrics.classification import MultilabelF1Score >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0.11, 0.22, 0.84], [0.73, 0.33, 0.92]]) >>> metric = MultilabelF1Score(num_labels=3) >>> metric(preds, target) tensor(0.5556) >>> metric = MultilabelF1Score(num_labels=3, average=None) >>> metric(preds, target) tensor([1.0000, 0.0000, 0.6667])
- Example (multidim tensors):
>>> from torchmetrics.classification import MultilabelF1Score >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> metric = MultilabelF1Score(num_labels=3, multidim_average='samplewise') >>> metric(preds, target) tensor([0.4444, 0.0000]) >>> metric = MultilabelF1Score(num_labels=3, multidim_average='samplewise', average=None) >>> metric(preds, target) tensor([[0.6667, 0.6667, 0.0000], [0.0000, 0.0000, 0.0000]])
Initializes internal Module state, shared by both nn.Module and ScriptModule.
Functional Interface¶
f1_score¶
- torchmetrics.functional.f1_score(preds, target, beta=1.0, average='micro', mdmc_average=None, ignore_index=None, num_classes=None, threshold=0.5, top_k=None, multiclass=None, task=None, num_labels=None, multidim_average='global', validate_args=True)[source]
F1 score.
Note
From v0.10 an
'binary_*','multiclass_*','multilabel_*'version now exist of each classification metric. Moving forward we recommend using these versions. This base metric will still work as it did prior to v0.10 until v0.11. From v0.11 the task argument introduced in this metric will be required and the general order of arguments may change, such that this metric will just function as an single entrypoint to calling the three specialized versions.Computes F1 metric. F1 metrics correspond to equally weighted average of the precision and recall scores.
Works with binary, multiclass, and multilabel data. Accepts probabilities or logits from a model output or integer class values in prediction. Works with multi-dimensional preds and target.
If preds and target are the same shape and preds is a float tensor, we use the
self.thresholdargument to convert into integer labels. This is the case for binary and multi-label probabilities or logits.If preds has an extra dimension as in the case of multi-class scores we perform an argmax on
dim=1.The reduction method (how the precision scores are aggregated) is controlled by the
averageparameter, and additionally by themdmc_averageparameter in the multi-dimensional multi-class case. Accepts all inputs listed in Input types.- Parameters
preds¶ (
Tensor) – Predictions from model (probabilities, logits or labels)average¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied. Should be one of the following:
'micro'[default]: Calculate the metric globally, across all samples and classes.'macro': Calculate the metric for each class separately, and average the metrics across classes (with equal weights for each class).'weighted': Calculate the metric for each class separately, and average the metrics across classes, weighting each class by its support (tp + fn).'none'orNone: Calculate the metric for each class separately, and return the metric for every class.'samples': Calculate the metric for each sample, and average the metrics across samples (with equal weights for each sample).
Note
What is considered a sample in the multi-dimensional multi-class case depends on the value of
mdmc_average.Note
If
'none'and a given class doesn’t occur in thepredsortarget, the value for the class will benan.mdmc_average¶ (
Optional[str]) –Defines how averaging is done for multi-dimensional multi-class inputs (on top of the
averageparameter). Should be one of the following:None[default]: Should be left unchanged if your data is not multi-dimensional multi-class.'samplewise': In this case, the statistics are computed separately for each sample on theNaxis, and then averaged over samples. The computation for each sample is done by treating the flattened extra axes...(see Input types) as theNdimension within the sample, and computing the metric for the sample based on that.'global': In this case theNand...dimensions of the inputs (see Input types) are flattened into a newN_Xsample axis, i.e. the inputs are treated as if they were(N_X, C). From here on theaverageparameter applies as usual.
ignore_index¶ (
Optional[int]) – Integer specifying a target class to ignore. If given, this class index does not contribute to the returned score, regardless of reduction method. If an index is ignored, andaverage=Noneor'none', the score for the ignored class will be returned asnan.num_classes¶ (
Optional[int]) – Number of classes. Necessary for'macro','weighted'andNoneaverage methods.threshold¶ (
float) – Threshold for transforming probability or logit predictions to binary (0,1) predictions, in the case of binary or multi-label inputs. Default value of 0.5 corresponds to input being probabilities.Number of highest probability or logit score predictions considered to find the correct label, relevant only for (multi-dimensional) multi-class inputs. The default value (
None) will be interpreted as 1 for these inputs.Should be left at default (
None) for all other types of inputs.multiclass¶ (
Optional[bool]) – Used only in certain special cases, where you want to treat inputs as a different type than what they appear to be. See the parameter’s documentation section for a more detailed explanation and examples.
- Return type
- Returns
The shape of the returned tensor depends on the
averageparameterIf
average in ['micro', 'macro', 'weighted', 'samples'], a one-element tensor will be returnedIf
average in ['none', None], the shape will be(C,), whereCstands for the number of classes
Example
>>> from torchmetrics.functional import f1_score >>> target = torch.tensor([0, 1, 2, 0, 1, 2]) >>> preds = torch.tensor([0, 2, 1, 0, 0, 1]) >>> f1_score(preds, target, num_classes=3) tensor(0.3333)
binary_f1_score¶
- torchmetrics.functional.classification.binary_f1_score(preds, target, threshold=0.5, multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes F-1 score for binary tasks:
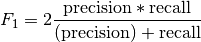
Accepts the following input tensors:
preds(int or float tensor):(N, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary {0,1} predictionsmultidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Return type
- Returns
If
multidim_averageis set toglobal, the metric returns a scalar value. Ifmultidim_averageis set tosamplewise, the metric returns(N,)vector consisting of a scalar value per sample.
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import binary_f1_score >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0, 0, 1, 1, 0, 1]) >>> binary_f1_score(preds, target) tensor(0.6667)
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import binary_f1_score >>> target = torch.tensor([0, 1, 0, 1, 0, 1]) >>> preds = torch.tensor([0.11, 0.22, 0.84, 0.73, 0.33, 0.92]) >>> binary_f1_score(preds, target) tensor(0.6667)
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import binary_f1_score >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> binary_f1_score(preds, target, multidim_average='samplewise') tensor([0.5000, 0.0000])
multiclass_f1_score¶
- torchmetrics.functional.classification.multiclass_f1_score(preds, target, num_classes, average='macro', top_k=1, multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes F-1 score for multiclass tasks:
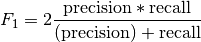
Accepts the following input tensors:
preds:(N, ...)(int tensor) or(N, C, ..)(float tensor). If preds is a floating point we applytorch.argmaxalong theCdimension to automatically convert probabilities/logits into an int tensor.target(int tensor):(N, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
num_classes¶ (
int) – Integer specifing the number of classesaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
top_k¶ (
int) – Number of highest probability or logit score predictions considered to find the correct label. Only works whenpredscontain probabilities/logits.multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import multiclass_f1_score >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([2, 1, 0, 1]) >>> multiclass_f1_score(preds, target, num_classes=3) tensor(0.7778) >>> multiclass_f1_score(preds, target, num_classes=3, average=None) tensor([0.6667, 0.6667, 1.0000])
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import multiclass_f1_score >>> target = torch.tensor([2, 1, 0, 0]) >>> preds = torch.tensor([ ... [0.16, 0.26, 0.58], ... [0.22, 0.61, 0.17], ... [0.71, 0.09, 0.20], ... [0.05, 0.82, 0.13], ... ]) >>> multiclass_f1_score(preds, target, num_classes=3) tensor(0.7778) >>> multiclass_f1_score(preds, target, num_classes=3, average=None) tensor([0.6667, 0.6667, 1.0000])
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import multiclass_f1_score >>> target = torch.tensor([[[0, 1], [2, 1], [0, 2]], [[1, 1], [2, 0], [1, 2]]]) >>> preds = torch.tensor([[[0, 2], [2, 0], [0, 1]], [[2, 2], [2, 1], [1, 0]]]) >>> multiclass_f1_score(preds, target, num_classes=3, multidim_average='samplewise') tensor([0.4333, 0.2667]) >>> multiclass_f1_score(preds, target, num_classes=3, multidim_average='samplewise', average=None) tensor([[0.8000, 0.0000, 0.5000], [0.0000, 0.4000, 0.4000]])
multilabel_f1_score¶
- torchmetrics.functional.classification.multilabel_f1_score(preds, target, num_labels, threshold=0.5, average='macro', multidim_average='global', ignore_index=None, validate_args=True)[source]
Computes F-1 score for multilabel tasks:
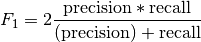
Accepts the following input tensors:
preds(int or float tensor):(N, C, ...). If preds is a floating point tensor with values outside [0,1] range we consider the input to be logits and will auto apply sigmoid per element. Addtionally, we convert to int tensor with thresholding using the value inthreshold.target(int tensor):(N, C, ...)
The influence of the additional dimension
...(if present) will be determined by the multidim_average argument.- Parameters
threshold¶ (
float) – Threshold for transforming probability to binary (0,1) predictionsaverage¶ (
Optional[Literal[‘micro’, ‘macro’, ‘weighted’, ‘none’]]) –Defines the reduction that is applied over labels. Should be one of the following:
micro: Sum statistics over all labelsmacro: Calculate statistics for each label and average themweighted: Calculates statistics for each label and computes weighted average using their support"none"orNone: Calculates statistic for each label and applies no reduction
multidim_average¶ (
Literal[‘global’, ‘samplewise’]) –Defines how additionally dimensions
...should be handled. Should be one of the following:global: Additional dimensions are flatted along the batch dimensionsamplewise: Statistic will be calculated independently for each sample on theNaxis. The statistics in this case are calculated over the additional dimensions.
ignore_index¶ (
Optional[int]) – Specifies a target value that is ignored and does not contribute to the metric calculationvalidate_args¶ (
bool) – bool indicating if input arguments and tensors should be validated for correctness. Set toFalsefor faster computations.
- Returns
If
multidim_averageis set toglobal:If
average='micro'/'macro'/'weighted', the output will be a scalar tensorIf
average=None/'none', the shape will be(C,)
If
multidim_averageis set tosamplewise:If
average='micro'/'macro'/'weighted', the shape will be(N,)If
average=None/'none', the shape will be(N, C)
- Return type
The returned shape depends on the
averageandmultidim_averagearguments
- Example (preds is int tensor):
>>> from torchmetrics.functional.classification import multilabel_f1_score >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0, 0, 1], [1, 0, 1]]) >>> multilabel_f1_score(preds, target, num_labels=3) tensor(0.5556) >>> multilabel_f1_score(preds, target, num_labels=3, average=None) tensor([1.0000, 0.0000, 0.6667])
- Example (preds is float tensor):
>>> from torchmetrics.functional.classification import multilabel_f1_score >>> target = torch.tensor([[0, 1, 0], [1, 0, 1]]) >>> preds = torch.tensor([[0.11, 0.22, 0.84], [0.73, 0.33, 0.92]]) >>> multilabel_f1_score(preds, target, num_labels=3) tensor(0.5556) >>> multilabel_f1_score(preds, target, num_labels=3, average=None) tensor([1.0000, 0.0000, 0.6667])
- Example (multidim tensors):
>>> from torchmetrics.functional.classification import multilabel_f1_score >>> target = torch.tensor([[[0, 1], [1, 0], [0, 1]], [[1, 1], [0, 0], [1, 0]]]) >>> preds = torch.tensor( ... [ ... [[0.59, 0.91], [0.91, 0.99], [0.63, 0.04]], ... [[0.38, 0.04], [0.86, 0.780], [0.45, 0.37]], ... ] ... ) >>> multilabel_f1_score(preds, target, num_labels=3, multidim_average='samplewise') tensor([0.4444, 0.0000]) >>> multilabel_f1_score(preds, target, num_labels=3, multidim_average='samplewise', average=None) tensor([[0.6667, 0.6667, 0.0000], [0.0000, 0.0000, 0.0000]])
